research
We have a broad range of research interests, spanning multiple domains and incorporating both computational and experimental techniques. Below are some current active areas of research.
Network Pharmacology & Precision Oncology
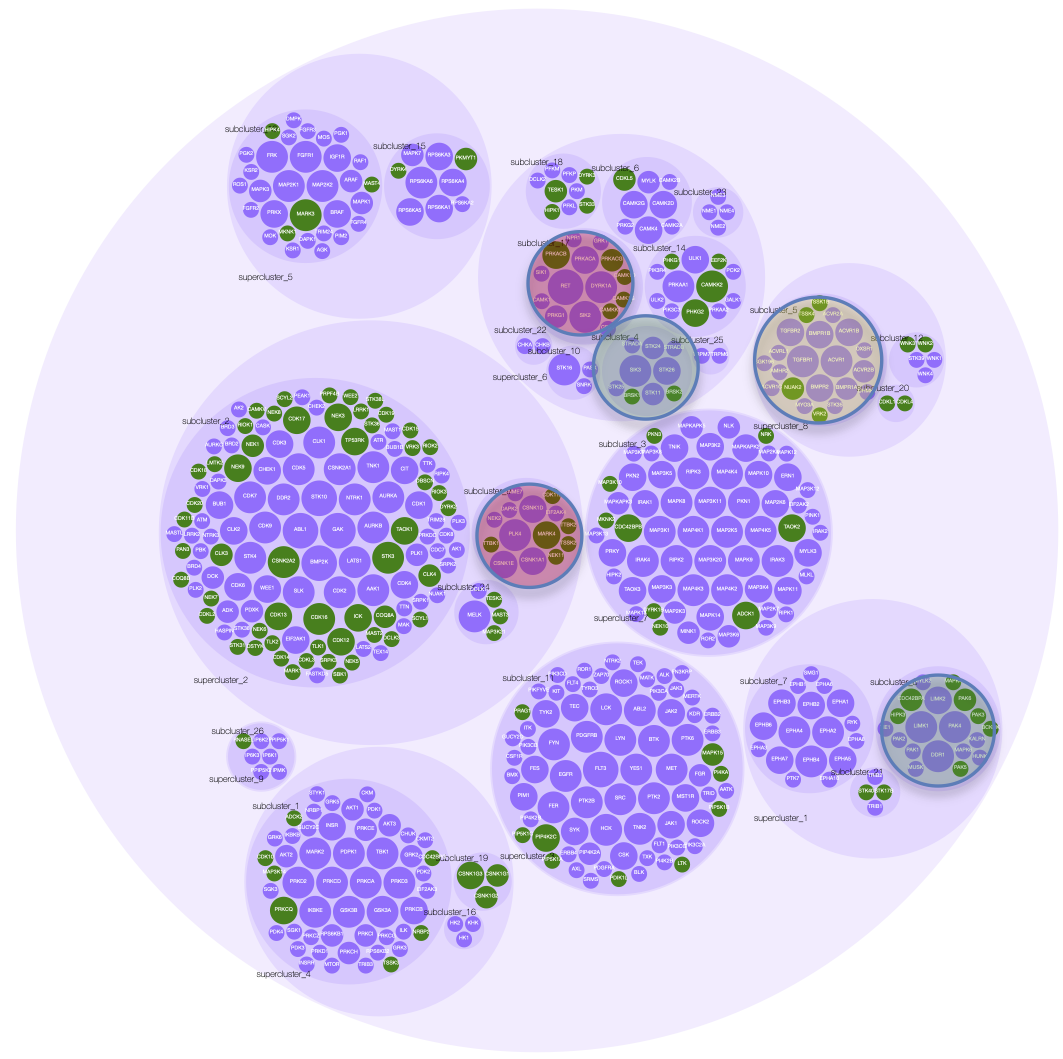
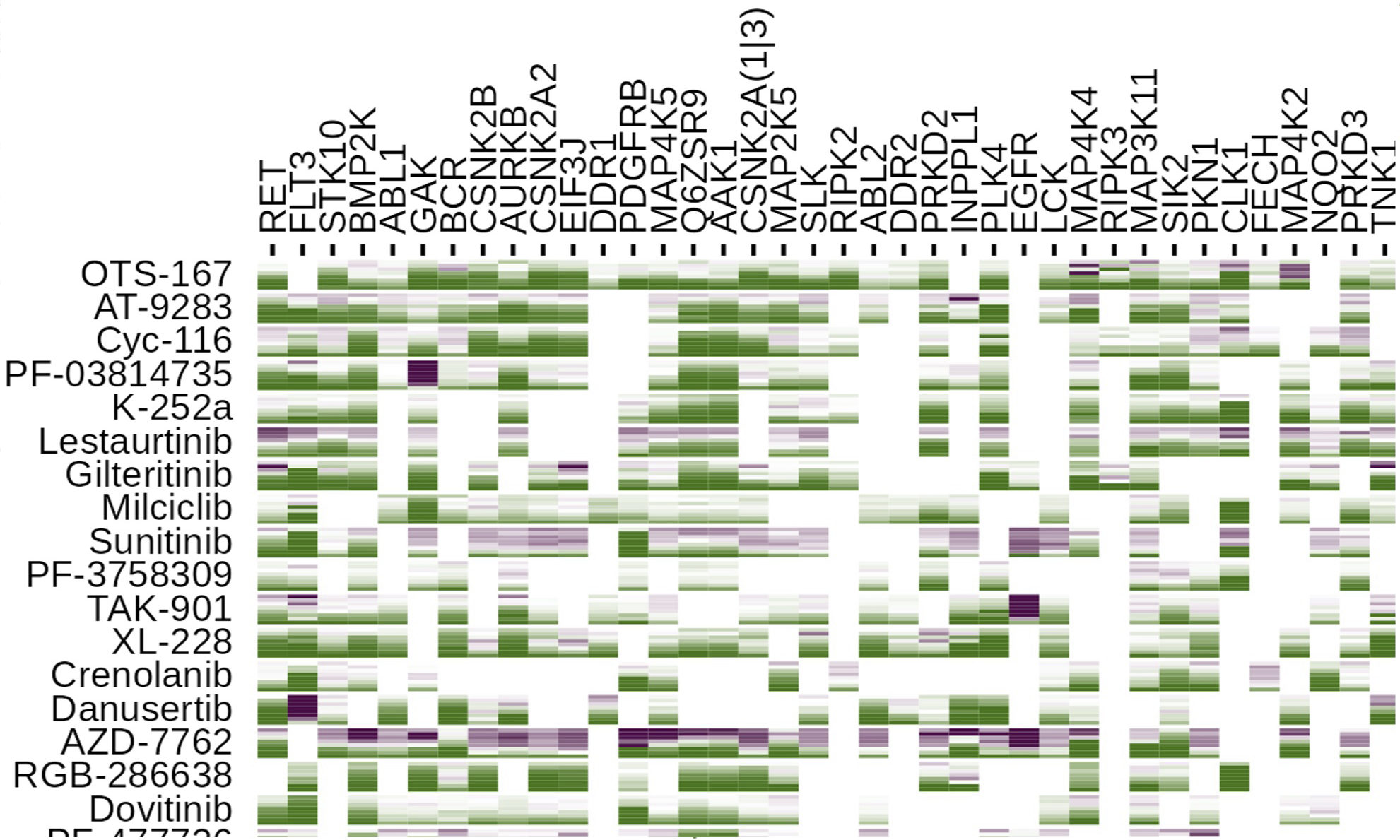
The kinome—the complete network of protein kinases in cells—plays a central role in cellular signaling and is frequently dysregulated in cancer and other diseases. Our laboratory combines proteomics, computational modeling, and systems biology approaches to map kinome architecture and predict responses to targeted therapies. We focus particularly on understudied "dark kinases" with therapeutic potential. A major emphasis is developing computational frameworks for rational design of combination therapies in cancer, integrating multi-omic data to predict which drug combinations will be most effective for individual patients. This work spans breast cancer, pancreatic cancer, and other solid tumors, with the goal of moving beyond trial-and-error approaches toward mechanism-based precision medicine.
Translational AI for Clinical Decision Support
Machine learning and AI offer transformative potential for healthcare, but translating these technologies into clinical practice remains challenging. Our group develops and validates AI models that augment clinical decision-making across the care continuum—from diagnosis to treatment planning to outcome prediction. Current applications include predictive models for surgical complications, cancer treatment response, margin assessment in oncologic surgery, and automated analysis of medical imaging. A core principle of our work is developing interpretable, clinically actionable models that can be prospectively validated and integrated into existing clinical workflows, with particular attention to issues of bias, generalizability, and regulatory pathways.
Computational Systems Biology & Drug Discovery
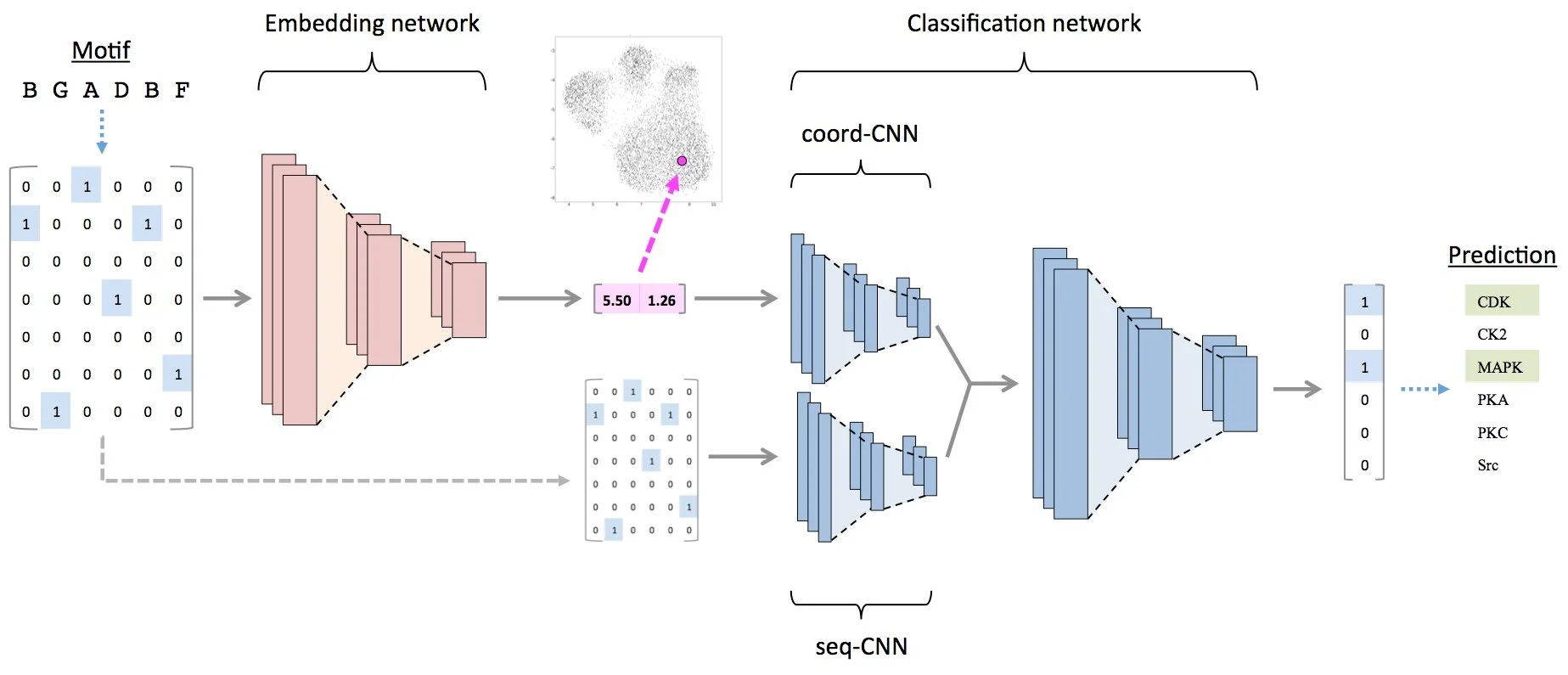
Understanding biological systems requires integrating diverse data types—genomics, proteomics, metabolomics—into coherent mechanistic frameworks. We develop computational methods to infer network structure, predict perturbation responses, and identify therapeutic vulnerabilities. This includes work on kinase-substrate prediction (EMBER), molecular representation learning (SALSA), and integration of chemical and biological networks. Our systems biology approaches span cancer biology, metabolic regulation, and host-pathogen interactions, with the ultimate goal of accelerating target identification and therapeutic development for diseases with unmet medical need.